ODEs with discontinuous forcing functions
September 01, 2011 at 01:56 PM | categories: odes | View Comments
ODEs with discontinuous forcing functions
John KitchinContents
Problem statement
Adapted from http://archives.math.utk.edu/ICTCM/VOL18/S046/paper.pdf A mixing tank initially contains 300 g of salt mixed into 1000 L of water. at t=0 min, a solution of 4 g/L salt enters the tank at 6 L/min. at t=10 min, the solution is changed to 2 g/L salt, still entering at 6 L/min. The tank is well stirred, and the tank solution leaves at a rate of 6 L/min. Plot the concentration of salt (g/L) in the tank as a function of time.Solution
A mass balance on the salt in the tank leads to this differential equation: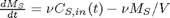 with the initial condition that
with the initial condition that 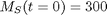 . The wrinkle is that
. The wrinkle is that
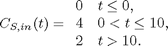
function main
tspan = [0,15]; % min Mo = 300; % gm salt [t,M] = ode45(@mass_balance,tspan,Mo); V = 1000; % L plot(t,M/V,'b.-') xlabel('Time (min)') ylabel('Salt concentration (g/L)')
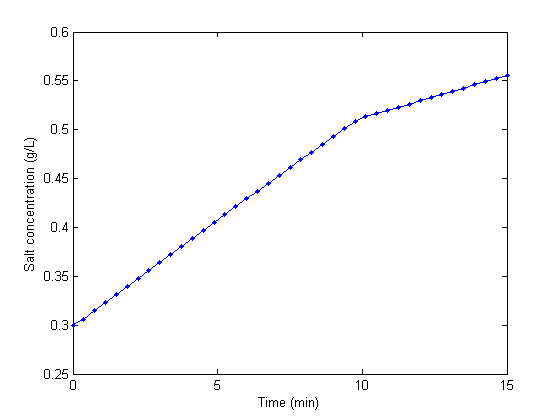 You can see the discontinuity in the salt concentration at 10 minutes due to the discontinous change in the entering salt concentration.
You can see the discontinuity in the salt concentration at 10 minutes due to the discontinous change in the entering salt concentration.
'done'
ans = donediscontinuous feed concentration function
function Cs = Cs_in(t)
if t < 0 Cs = 0; % g/L elseif t > 0 && t <= 10 Cs = 4; % g/L else Cs = 2; % g/L end
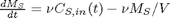
function dMsdt = mass_balance(t,Ms)
V = 1000; % L nu = 6; % L/min dMsdt = nu*Cs_in(t) - nu*Ms/V;
% categories: ODEs
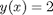 . In a numerical solution to an ODE we get a vector of independent variable values, and the corresponding function values at those values. To solve for a particular function value we need a different approach. This post will show one way to do that in Matlab.
. In a numerical solution to an ODE we get a vector of independent variable values, and the corresponding function values at those values. To solve for a particular function value we need a different approach. This post will show one way to do that in Matlab.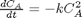 with the initial condition
with the initial condition 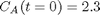 mol/L and
mol/L and 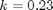 L/mol/s, compute the time it takes for
L/mol/s, compute the time it takes for 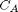 to be reduced to 1 mol/L.
to be reduced to 1 mol/L.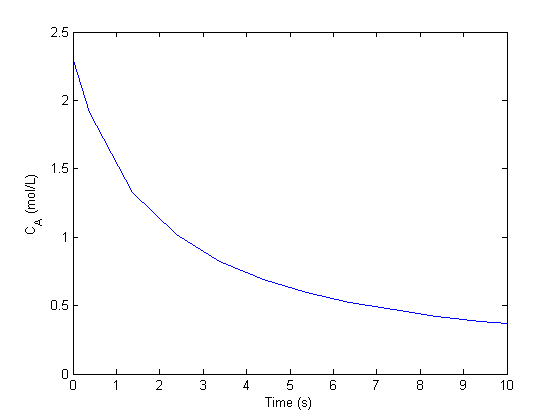
 where
where 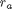 is the rate law. Suppose we know the reactor volume is 100 L, the inlet molar flow of A is 1 mol/L, the volumetric flow is 10 L/min, and
is the rate law. Suppose we know the reactor volume is 100 L, the inlet molar flow of A is 1 mol/L, the volumetric flow is 10 L/min, and  , with
, with 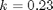 1/min. What is the exit molar flow rate? We need to solve the following equation:
1/min. What is the exit molar flow rate? We need to solve the following equation: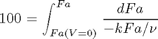
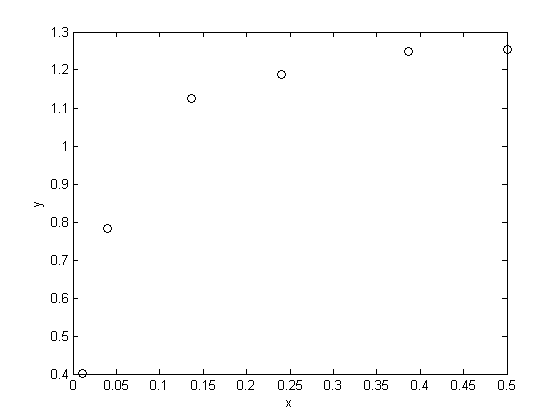
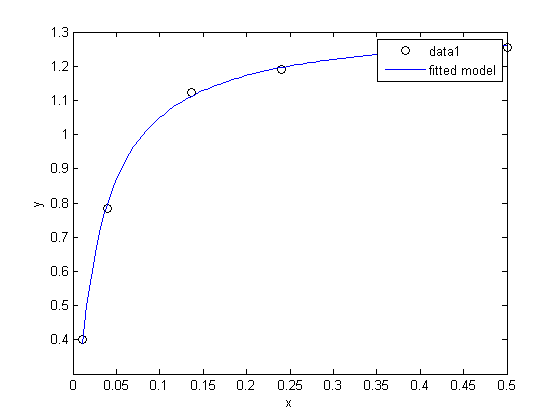
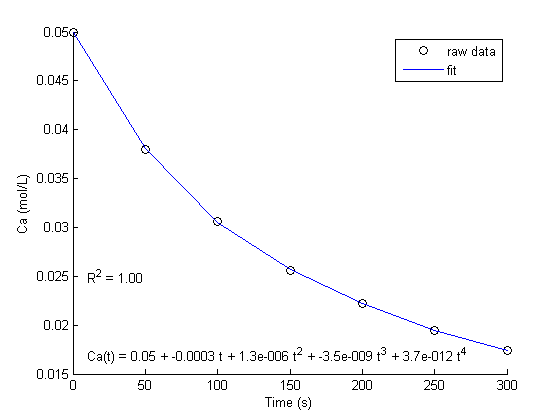
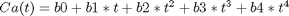 fit to the data in the least squares sense. We can write this in a linear algebra form as: T*b = Ca where T is a matrix of columns [1 t t^2 t^3 t^4], and b is a column vector of [b0 b1 b2 b3 b4]'. We want to solve for the b vector.
fit to the data in the least squares sense. We can write this in a linear algebra form as: T*b = Ca where T is a matrix of columns [1 t t^2 t^3 t^4], and b is a column vector of [b0 b1 b2 b3 b4]'. We want to solve for the b vector.